Thinking a pigeon is a bird of prey, because it’s perched in a tree, is easily my funniest iNaturalist computer vision fail yet.
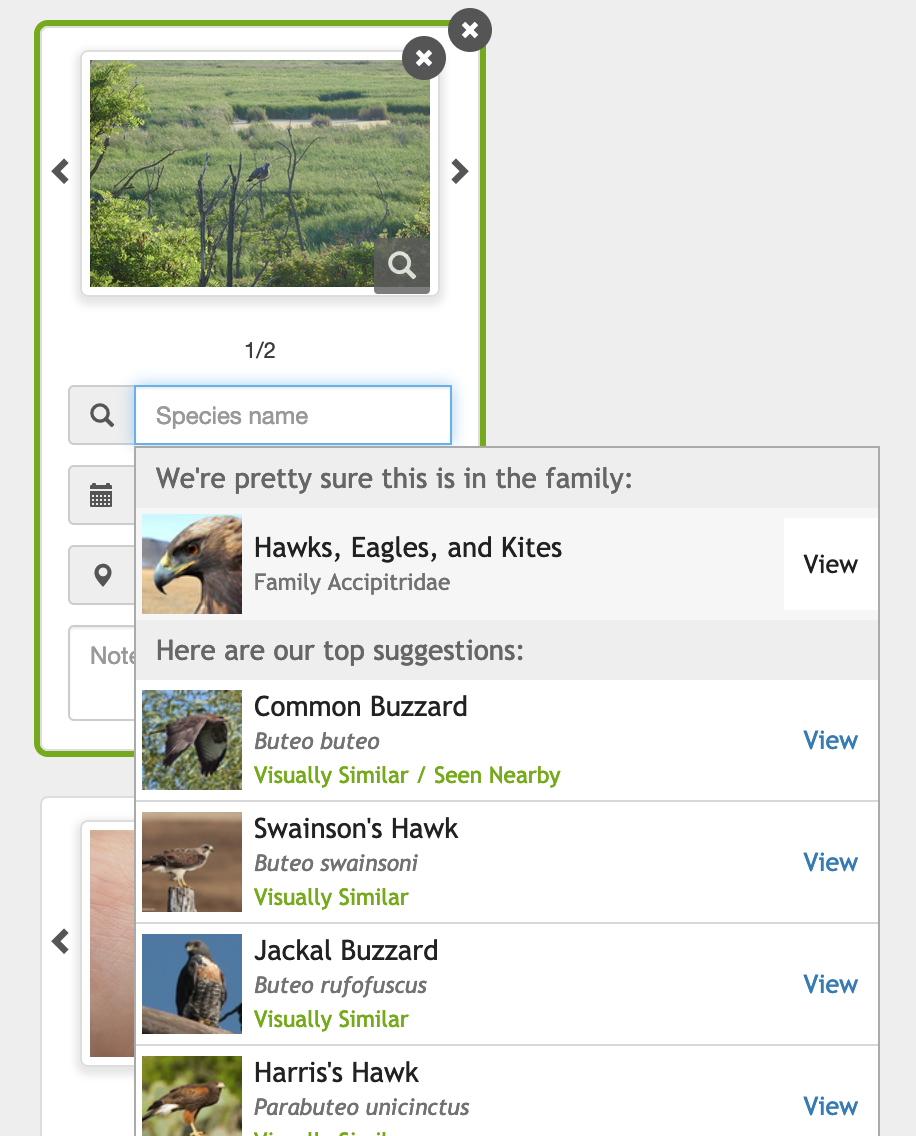

Thinking a pigeon is a bird of prey, because it’s perched in a tree, is easily my funniest iNaturalist computer vision fail yet.


Here’s a python snippet for analysing an iNaturalist export file and exporting an HTML-formatted list of species which only have observations from a single person (e.g. this list for the CNC Wien 2021)
# coding: utf-8
import argparse
import pandas as pd
"""
Find which species in an iNaturalist export only have observations from a single observer.
Get an export from here: https://www.inaturalist.org/observations/export with a query such
as quality_grade=research&identifications=any&rank=species&projects[]=92926 and at least the
following columns: taxon_id, scientific_name, common_name, user_login
Download it, extract the CSV, then run this script with the file name as its argument. It will
output basic stats formatted as HTML.
The only external module required is pandas.
Example usage:
py uniquely_observed_species.py wien_cnc_2021.csv > wien_cnc_2021_results.html
If you provide the --project-id (-p) argument, the taxa links in the output list will link to
a list of observations of that taxa within that project. Otherwise, they default to linking
to the taxa page.
If a quality_grade column is included, non-research-grade observations will be included in the
analysis. Uniquely observed species with no research-grade observations will be marked. Species
which were observed by multiple people, only one of which has research-grade observation(s) will
also be marked.
By Barnaby Walters waterpigs.co.uk
"""
if __name__ == "__main__":
parser = argparse.ArgumentParser(description='Given an iNaturalist observation export, find species which were only observed by a single person.')
parser.add_argument('export_file')
parser.add_argument('-p', '--project-id', dest='project_id', default=None)
args = parser.parse_args()
uniquely_observed_species = {}
df = pd.read_csv(args.export_file)
# If quality_grade isn’t given, assume that the export contains only RG observations.
if 'quality_grade' not in df.columns:
df.loc[:, 'quality_grade'] = 'research'
# Filter out casual observations.
df = df.query('quality_grade != "casual"')
# Create a local species reference from the dataframe.
species = df.loc[:, ('taxon_id', 'scientific_name', 'common_name')].drop_duplicates()
species = species.set_index(species.loc[:, 'taxon_id'])
for tid in species.index:
observers = df.query('taxon_id == @tid').loc[:, 'user_login'].drop_duplicates()
research_grade_observers = df.query('taxon_id == @tid and quality_grade == "research"').loc[:, 'user_login'].drop_duplicates()
if observers.shape[0] == 1:
# Only one person made any observations of this species.
observer = observers.squeeze()
if observer not in uniquely_observed_species:
uniquely_observed_species[observer] = []
uniquely_observed_species[observer].append({
'id': tid,
'has_research_grade': (not research_grade_observers.empty),
'num_other_observers': 0
})
elif research_grade_observers.shape[0] == 1:
# Multiple people observed the species, but only one person has research-grade observation(s).
rg_observer = research_grade_observers.squeeze()
if rg_observer not in uniquely_observed_species:
uniquely_observed_species[rg_observer] = []
uniquely_observed_species[rg_observer].append({
'id': tid,
'has_research_grade': True,
'num_other_observers': observers.shape[0] - 1
})
# Sort observers by number of unique species.
sorted_observations = sorted(uniquely_observed_species.items(), key=lambda t: len(t[1]), reverse=True)
print(f"<p>{sum([len(t) for _, t in sorted_observations])} taxa uniquely observed by {len(sorted_observations)} observers.</p>")
print('<p>')
for observer, _ in sorted_observations:
print(f"@{observer} ", end='')
print('</p>')
print('<p><b>bold</b> species are ones for which the given observer has one or more research-grade observations.</p>')
print('<p>If only one person has RG observations of a species, but other people have observations which need ID, the number of needs-ID observers are indicated in parentheses.')
for observer, taxa in sorted_observations:
print(f"""\n\n<p><a href="https://www.inaturalist.org/people/{observer}">@{observer}</a> ({len(taxa)} taxa):</p><ul>""")
for tobv in sorted(taxa, key=lambda t: species.loc[t['id']]['scientific_name']):
tid = tobv['id']
t = species.loc[tid]
if args.project_id:
taxa_url = f"https://www.inaturalist.org/observations?taxon_id={tid}&project_id={args.project_id}"
else:
taxa_url = f'https://www.inaturalist.org/taxa/{tid}'
rgb, rge = ('<b>', '</b>') if tobv.get('has_research_grade') else ('', '')
others = f" ({tobv.get('num_other_observers', 0)})" if tobv.get('num_other_observers', 0) > 0 else ''
if not pd.isnull(t['common_name']):
print(f"""<li><a href="{taxa_url}">{rgb}<i>{t['scientific_name']}</i> ({t['common_name']}){rge}{others}</a></li>""")
else:
print(f"""<li><a href="{taxa_url}">{rgb}<i>{t['scientific_name']}</i>{rge}{others}</a></li>""")
print("</ul>")