Another breakthrough in the increasingly relevant field of linguistic cryptozoology
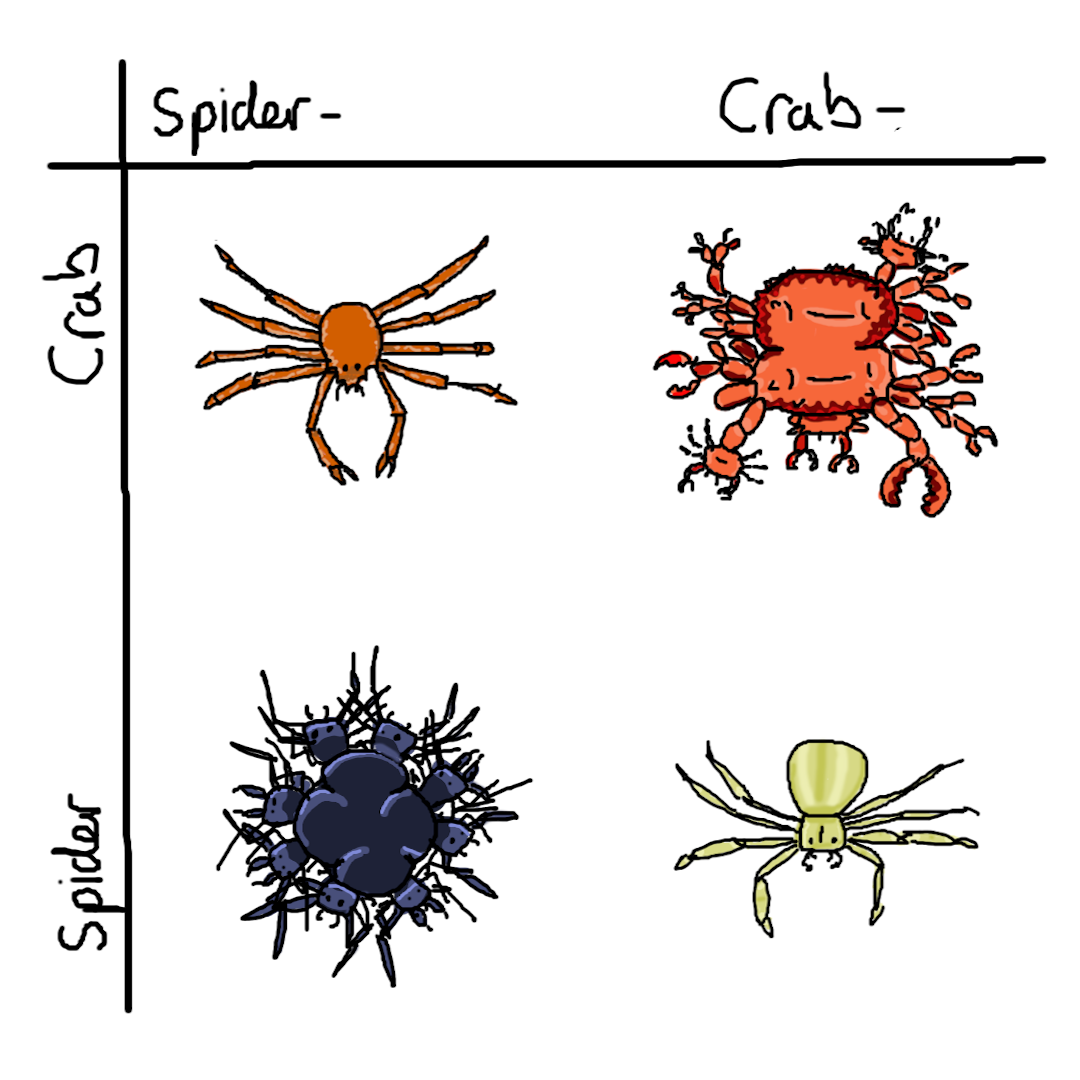
(previously: most weasel)
Another breakthrough in the increasingly relevant field of linguistic cryptozoology

(previously: most weasel)
That first one was definitely the funniest, but the some of the other bowl-demons are pretty great too.


There’s a bit more detail about exactly what’s going on in this article.
I’m surprised how timeless and appealing these little drawings are. They avoid a lot of the foibles which make a lot of late antique/medieval art look dated, and the exaggerated proportions, noodly limbs and googly eyes wouldn’t be out of place in a webcomic or cartoon.
I also love that these bowls refer to themselves as “amulets”. Next time someone describes something as an “amulet” in a novel or TTRPG I’m definitely going to imagine it as an inverted bowl with a googly-eyed stick figure demon on.
toad opera



bonus frog

I’ve wanted to see an Alpine Rosalia (Rosalia alpina) ever since I first heard about them, and finally managed to spot two today! Very impressive beetles, lots of fun to watch them move, and they make a cute little scratching sound when disturbed.


Recently found this fun local Austrian legend about @apocrypals’ favourite 12 foot tall werewolf saint.



An ominous notification to come home to

#TIL that VLC player has a built in compressor, in Window → Audio Effects, of all places. A boon for poorly mixed podcast listening!
Thinking a pigeon is a bird of prey, because it’s perched in a tree, is easily my funniest iNaturalist computer vision fail yet.

New #indieweb libraries: taproot/micropub-adapter and taproot/indieauth!
Finally put the finishing touches on these two closely-related libraries, which make it quick and easy to add Micropub and IndieAuth support to any PHP app which uses PSR-7.
Feedback appreciated, either as replies, GH issues, or at indieweb.org/discuss
Extremely cursed UI at neverwinter.fandom.com/wiki/Ring_of_Warding
PHPUnit’s HTML code coverage reports don’t play nicely with GitHub pages “main branch /docs folder” by default, as they store CSS, JS and icon assets in folders prefixed with underscores.
Here’s a little bash script to run tests with code coverage enabled, then move the assets around:
rm -rf docs/coverage/
XDEBUG_MODE=coverage ./vendor/bin/phpunit tests --coverage-filter src --coverage-html docs/coverage
mv docs/coverage/_css docs/coverage/phpunit_css
mv docs/coverage/_icons docs/coverage/phpunit_icons
mv docs/coverage/_js docs/coverage/phpunit_js
grep -rl _css docs/coverage | xargs sed -i "" -e 's/_css/phpunit_css/g'
grep -rl _icons docs/coverage | xargs sed -i "" -e 's/_icons/phpunit_icons/g'
grep -rl _js docs/coverage | xargs sed -i "" -e 's/_js/phpunit_js/g'
That allows you to use GitHub pages to show code coverage reports as well as docs, as I’m doing for taproot/indieauth.
Some more hamster activity today! Anyone have any idea what the behaviour at 0:57 is? Looks like it’s trying to flatten grass, but I imagine it’d rather rest in its hole than on the surface.
Update: based on discussion in this thread, looks like it’s probably scent-marking after a territorial dispute with another male.
If the micrometer is to be believed, these maple shavings are 0.015mm thick. Amazing what’s possible with sharp tools!

Ridiculous amounts of hamster activity at the moment. I must have seen nearly 100 of them in an hour. I also heard them vocalise for the first time, a sort of guttural hissing sound.


#TIL there’s a species of moth called Wockia chewbacca. Someone must have had fun naming that one
Tip for anyone using greg to download podcasts: this config setting forces the downloaded files to be in chronological order, and strips any query parameters from the end of their name:
downloadhandler = wget {link} -O {directory}/{date}_{filename}
Here’s a python snippet for analysing an iNaturalist export file and exporting an HTML-formatted list of species which only have observations from a single person (e.g. this list for the CNC Wien 2021)
# coding: utf-8
import argparse
import pandas as pd
"""
Find which species in an iNaturalist export only have observations from a single observer.
Get an export from here: https://www.inaturalist.org/observations/export with a query such
as quality_grade=research&identifications=any&rank=species&projects[]=92926 and at least the
following columns: taxon_id, scientific_name, common_name, user_login
Download it, extract the CSV, then run this script with the file name as its argument. It will
output basic stats formatted as HTML.
The only external module required is pandas.
Example usage:
py uniquely_observed_species.py wien_cnc_2021.csv > wien_cnc_2021_results.html
If you provide the --project-id (-p) argument, the taxa links in the output list will link to
a list of observations of that taxa within that project. Otherwise, they default to linking
to the taxa page.
If a quality_grade column is included, non-research-grade observations will be included in the
analysis. Uniquely observed species with no research-grade observations will be marked. Species
which were observed by multiple people, only one of which has research-grade observation(s) will
also be marked.
By Barnaby Walters waterpigs.co.uk
"""
if __name__ == "__main__":
parser = argparse.ArgumentParser(description='Given an iNaturalist observation export, find species which were only observed by a single person.')
parser.add_argument('export_file')
parser.add_argument('-p', '--project-id', dest='project_id', default=None)
args = parser.parse_args()
uniquely_observed_species = {}
df = pd.read_csv(args.export_file)
# If quality_grade isn’t given, assume that the export contains only RG observations.
if 'quality_grade' not in df.columns:
df.loc[:, 'quality_grade'] = 'research'
# Filter out casual observations.
df = df.query('quality_grade != "casual"')
# Create a local species reference from the dataframe.
species = df.loc[:, ('taxon_id', 'scientific_name', 'common_name')].drop_duplicates()
species = species.set_index(species.loc[:, 'taxon_id'])
for tid in species.index:
observers = df.query('taxon_id == @tid').loc[:, 'user_login'].drop_duplicates()
research_grade_observers = df.query('taxon_id == @tid and quality_grade == "research"').loc[:, 'user_login'].drop_duplicates()
if observers.shape[0] == 1:
# Only one person made any observations of this species.
observer = observers.squeeze()
if observer not in uniquely_observed_species:
uniquely_observed_species[observer] = []
uniquely_observed_species[observer].append({
'id': tid,
'has_research_grade': (not research_grade_observers.empty),
'num_other_observers': 0
})
elif research_grade_observers.shape[0] == 1:
# Multiple people observed the species, but only one person has research-grade observation(s).
rg_observer = research_grade_observers.squeeze()
if rg_observer not in uniquely_observed_species:
uniquely_observed_species[rg_observer] = []
uniquely_observed_species[rg_observer].append({
'id': tid,
'has_research_grade': True,
'num_other_observers': observers.shape[0] - 1
})
# Sort observers by number of unique species.
sorted_observations = sorted(uniquely_observed_species.items(), key=lambda t: len(t[1]), reverse=True)
print(f"<p>{sum([len(t) for _, t in sorted_observations])} taxa uniquely observed by {len(sorted_observations)} observers.</p>")
print('<p>')
for observer, _ in sorted_observations:
print(f"@{observer} ", end='')
print('</p>')
print('<p><b>bold</b> species are ones for which the given observer has one or more research-grade observations.</p>')
print('<p>If only one person has RG observations of a species, but other people have observations which need ID, the number of needs-ID observers are indicated in parentheses.')
for observer, taxa in sorted_observations:
print(f"""\n\n<p><a href="https://www.inaturalist.org/people/{observer}">@{observer}</a> ({len(taxa)} taxa):</p><ul>""")
for tobv in sorted(taxa, key=lambda t: species.loc[t['id']]['scientific_name']):
tid = tobv['id']
t = species.loc[tid]
if args.project_id:
taxa_url = f"https://www.inaturalist.org/observations?taxon_id={tid}&project_id={args.project_id}"
else:
taxa_url = f'https://www.inaturalist.org/taxa/{tid}'
rgb, rge = ('<b>', '</b>') if tobv.get('has_research_grade') else ('', '')
others = f" ({tobv.get('num_other_observers', 0)})" if tobv.get('num_other_observers', 0) > 0 else ''
if not pd.isnull(t['common_name']):
print(f"""<li><a href="{taxa_url}">{rgb}<i>{t['scientific_name']}</i> ({t['common_name']}){rge}{others}</a></li>""")
else:
print(f"""<li><a href="{taxa_url}">{rgb}<i>{t['scientific_name']}</i>{rge}{others}</a></li>""")
print("</ul>")